Our
Research
Research at the Rabadan Lab
Our main scientific interests lie in modeling and understanding the dynamics of biological systems through the lens of genomics. We are a very interdisciplinary team of mathematicians, physicists, engineers, biologists, and medical doctors with a common goal of solving pressing medical problems. We are currently focusing our work on:
Cancer. Genomic technologies provide an extraordinary opportunity to identify mutations that contribute to the development of tumors. We are mapping the evolution of cancers and uncovering the mechanisms of response or lack of response to multiple therapies. We work with clinicians and experimentalists all around the world.
Infectious diseases. Evolution is a dynamic process that shapes genomes. Our team at Columbia is developing algorithms to analyze genomic data, with a view to understanding the molecular biology, population genetics, phylogeny, and epidemiology of viruses. We are interested in the emergence of infectious diseases, pandemics and uncovering the mechanisms of adaptation of viruses to humans.
Electronic Health Records. Clinical databases constitute a rich and complex source of raw data. We are using the power of statistics and computers to tease out important clinical patterns in these diverse, important datasets. Combining molecular and clinical data illuminates some of the mechanisms underlying complex diseases.
In particular, we develop mathematical, statistical, and computational approaches, which cover the analysis of high throughput data right through to the altogether more abstract identification of global patterns in evolutionary processes. Learn more about the Rabadan Lab and the three main global questions that we are addressing.
Research Projects
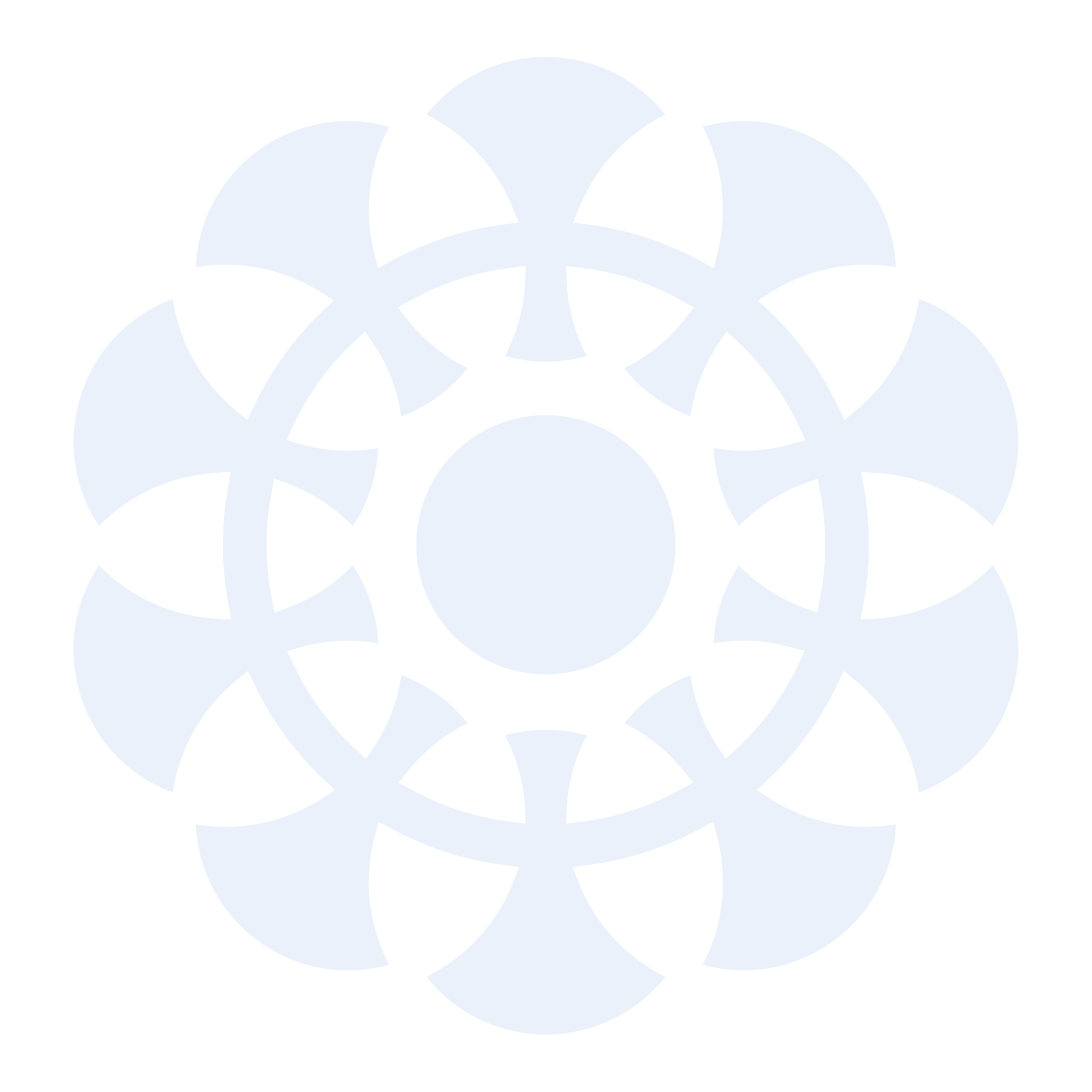
Genetic mechanisms of HLA-I loss and immune escape in diffuse large B cell lymphoma
Fifty percent of diffuse large B cell lymphoma (DLBCL) evade immune-surveillance via somatic genetic lesions abrogating the expression of the class I major histocompatibility complex (MHC-I) complex on the cell surface, thus preventing the presentation of tumor neoantigens to the immune system. The results herein significantly extend these findings by showing that an additional 40% of DLBCL cases, despite expressing MHC-I, carry monoallelic HLA-I genetic alterations that limit the repertoire of neoantigens for presentation to immune cells. Both MHC-I negative and MHC-I positive/monoallelically disrupted cases have significantly higher mutational load. Notably, homozygosis of HLA-I loci is significantly and preferentially enriched in the germline of DLBCL patients, suggesting a stepwise process by which limited neoantigen presentation is selected during DLBCL development.